Discovery is part of science. Although many experiments are hypothesis driven, the thrill of discovering something new motivates most scientists-whether that be discovering a new planet, a new medical treatment, or a new organism.
When we think of living things most of us picture the plants and animals around us-the ones we can see. But, the vast majority of living things that exist on our planet are invisible to the naked eye, and sadly not well characterized. Many microbial organisms can’t be grown in the laboratory-or at least we don’t know how to grow them yet. But DNA analysis can be used to identify new organisms and help us understand what role these organisms play in the ecosystem from which they are found. Many of the organisms we “see” are consumers of detritus, algae blooms, or are recyclers. These are in themselves part of the food web of crabs, fish and even birds that live in the estuarine environment.
Methods
We are using environmental metagenomics to comprehensively catalog the organisms living in the Inner Harbor and Chesapeake Bay. Our workflow consists of:
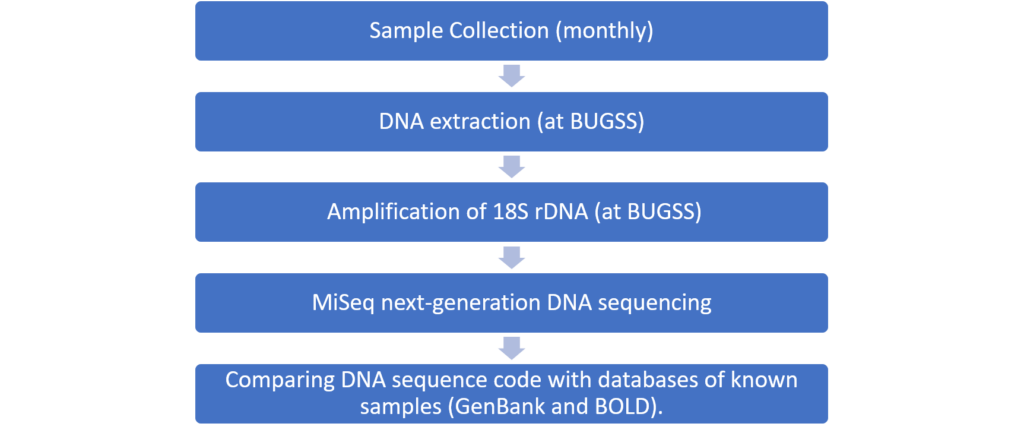
Results
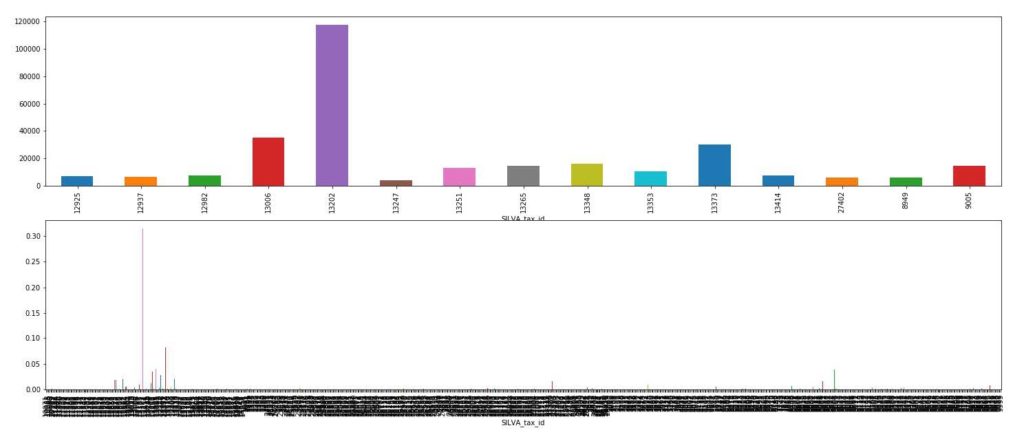
Over 200 putative species (operational taxonomic units) were identified in the DNA sequences. Most of them were seen only a few times (bottom chart). Fifteen species were present in more than 1% of the approximately 1 million sequence reads (top chart). Sequence code numbers listed under histograms correspond to the groups of animals shown at the top.

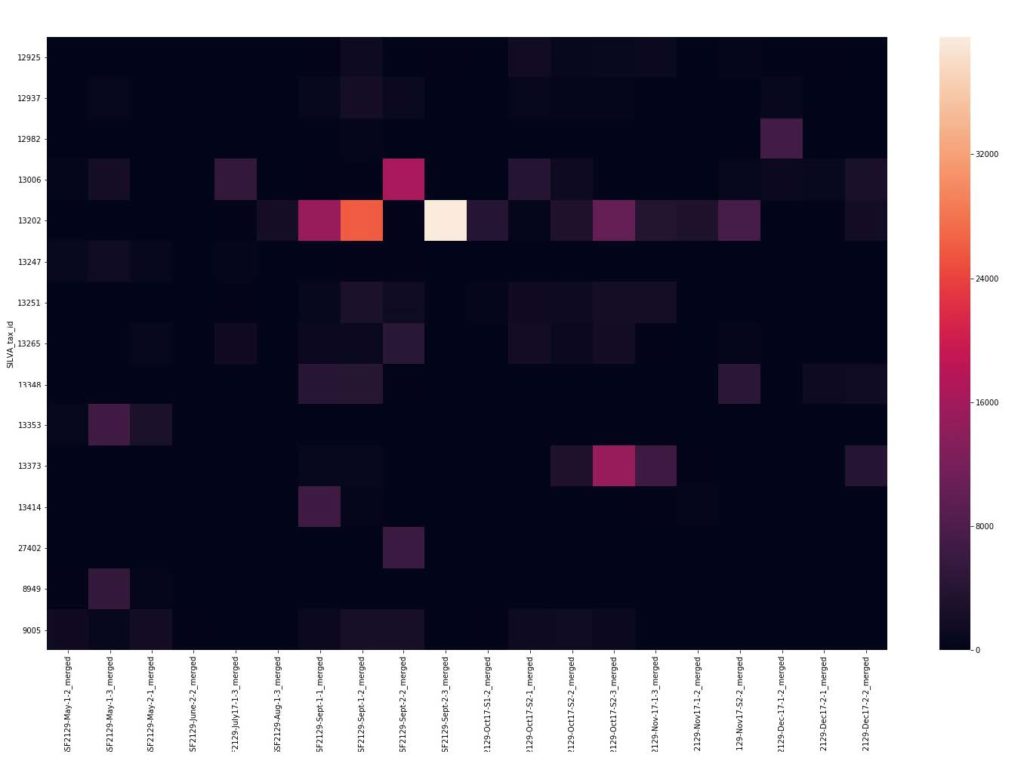
We also saw a tremendous variability in the species that were present over time. The heat map depicts the relative abundance of each organism (OTU) present at >1% abundance (Y-axis) at each sampling (X-axis).
When looking at the identity of these individual organisms, we can see that almost all species show fluctuation in their abundance across the seasons. The individual bar plots below show the sequence counts for each species in the restricted data set (>1% abundance).

Where are we going from here?
We still have more samples to analyze! We’ll be filling in the data with more dates and with samples from different years. Will we see the same trends year after year or will things change depending on the weather and other factors such as pollution that affect the Habor?
We also have some mysteries to solve. We see that many of the sequences we have recovered in this investigation are similar to, but not identical to, known groups of marine invertebrates. It’s probable that some of these are species that have not yet been sequenced. Some may not yet be recognized as distinct species, and some could be new to science! Could these organisms harbor unique enzymes that could be used medicinally, be used to degrade pollutants, or help us understand how nutrients cycle through the environment?
How can you help?
Become a part of the BUGSS/IMET Barcoding team! Help us to sequence more DNA, identify our unknown organisms, and maybe, just maybe, you’ll get to name one!
Email Lisa at lscheifele@bugssonline.org for info and to get involved!
We’ll have classes on Extracting DNA from Harbor samples and Analyzing DNA from Harbor samples coming up in fall 2019.
Thanks!

To our funders, the Chesapeake Bay Trust!